Systems Biology: Studying the World‘s Most Complex Dynamic Systems
By Ricardo Paxson, MathWorks and Kristen Zannella, MathWorks

The drug Artemisinin, made from the leaves of the sweet wormwood shrub, is a proven cure for malaria. At $2 per dose, however, it is far beyond the reach of the developing countries that need it most. A team of systems biologists solved this problem by designing a new organism that produces the same drug compound for $.02 a dose.
From medicine and environmental science to alternative fuel technology and agriculture, systems biologists are literally changing the world. Revolutionary new pharmaceuticals have moved to clinical trials in a fraction of the time taken by traditional methods. Systems biologists are not only accelerating the drug discovery process, they are also developing synthetic viruses to attack cancer cells, creating biosensors to detect arsenic in drinking water, developing biofuels, and designing algae that process carbon dioxide to reduce powerplant emissions.
By studying the relationships between the components that make up an organism, systems biologists build a systems-level understanding of how the biological world works. Like engineers, they solve problems by understanding systems and then applying that knowledge to control them. As a result, systems biology is not only a scientific discipline but also an engineering one.
Applying Engineering Techniques to Systems Biology
While the techniques used to build aircraft and automobiles, such as modeling, simulation, and computation, can be applied in systems biology, few research labs have used them successfully. This is partly because the researchers lack the necessary tools. It is also because most biological systems are much more complex than even the most sophisticated aircraft, and it takes a significant amount of reverse-engineering to gather enough information and insight to model them.
Faced with these obstacles, many systems biologists resort to more traditional methods, such as testing drug candidates on animals. To an engineer, this trial-and-error approach might seem equivalent to an aerospace company’s building multiple prototypes of planes to see which one flies best.
The cost, inefficiency, and potential risks of the trial-and-error approach are compelling more and more systems biologists to break through the obstacles and adopt engineering techniques and technology. Before testing a drug candidate on animals or humans, for example, they might first develop computational models of drug candidates and then run simulations to reject those with little chance of success and optimize the most promising ones.
Engineering Inroads
While modeling and simulation have yet to be universally adopted, three common engineering techniques are becoming widely used in systems biology: parameter estimation, simulation, and sensitivity analysis.
Engineers use parameter estimation to calibrate the response of a model to the observed outputs of a physical system. Instead of using educated guesses to adjust model parameters and initial conditions, they automatically compute these values using data gathered through test runs or experiments.
For example, a mechanical engineer designing a DC motor will include model parameters such as shaft inertia, viscous friction (damping), armature resistance, and armature inductance. While estimates for some of these values may be available from manufacturers, by using test results and parameter estimation, the engineer can find parameter values that enable the model response to accurately reflect the actual system.
Parameter estimation is a vital capability in systems biology because it enables researchers to generate approximate values for model parameters based on data gathered in experiments. In many cases, researchers know what species or molecular components must be present in the model or how species react with one another, but lack reliable estimates for model parameters such as reaction rates and concentrations. Often, researchers lack these values because the wet-bench experiments needed to determine them directly are too difficult or costly, or there is no published data on the parameters. Parameter estimation lets them calculate these values, enabling simulation and analysis.
Engineers use simulation to observe the system in action, change its inputs, parameters, and components, and analyze the results computationally.
Most engineering simulations are deterministic: Motor, circuits, and control systems must all provide the same outputs for a given set of inputs for each simulation run. Biological simulations, on the other hand, must incorporate the innate randomness of nature. For example, reactions occur with a certain probability, and a compound that is bound in one simulation might not be bound in the next. To account for this randomness, systems biologists use Monte Carlo techniques and stochastic simulations.
Sensitivity analysis enables engineers to determine which components of the model have the greatest effect on its output. For example, aerospace engineers use computational fluid dynamics on the geometry of an airplane wing to reduce drag. They perform sensitivity analysis on each point along the wing to discover which change has the most effect on the drag.
In systems biology, sensitivity analysis provides a computational mechanism to determine which parameters are most important under a specific set of conditions. In a model with 200 species and 100 different parameters, being able to determine which species and parameters most effect the desired output can eliminate fruitless avenues of research and enable scientists to focus wet-bench experiments on the most promising candidates.
Obstacles to Widespread Adoption
While these techniques have great potential in systems biology, biologists have not yet applied them as efficiently as engineers in traditional disciplines—both because of the complexity of biological systems and because systems biology research requires contributions from a diverse group of researchers. Modelers understand the computational approach and the mathematics behind it, while the scientists know the underlying biology. The two groups frequently use a different vocabulary and work with different concepts and tools.
An Environment for Systems Biologists
An engineer might wonder, why not use Simulink, the MathWorks platformfor simulation and Model-Based Design? While it is clear that engineers and biologists benefit from the same kinds of modeling and simulation techniques, it is equally clear that the two groups cannot use the same tools. Simulink was built for engineers, and was designed with an engineering look and feel—one that does not resonate with biologists.
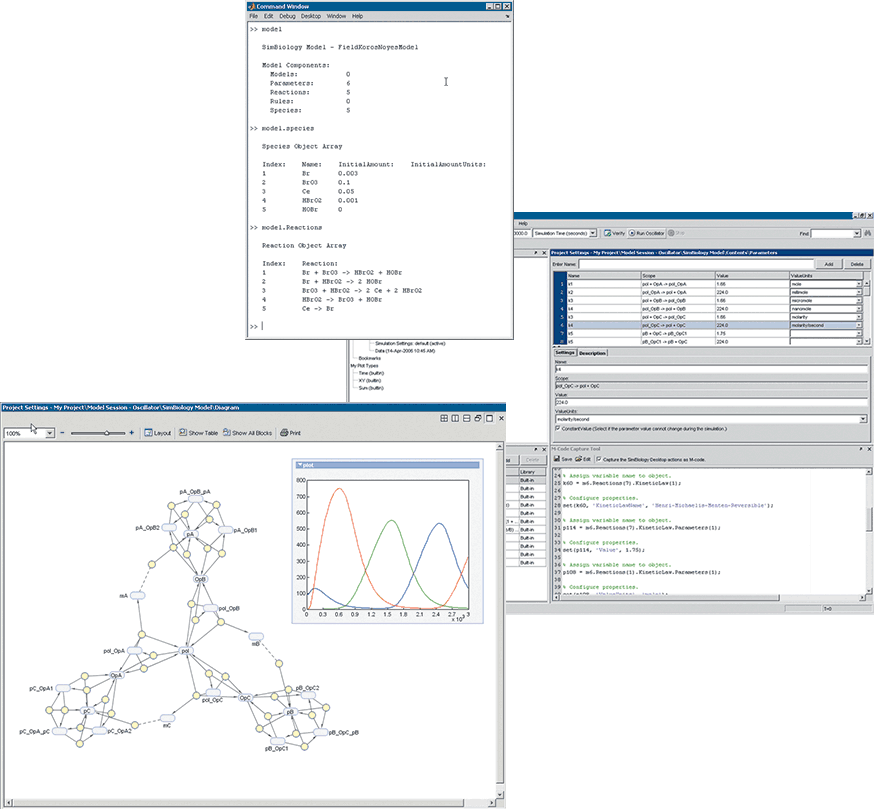
It was in response to these requirements that The MathWorks developed SimBiology. Like Simulink, SimBiology builds on the MATLAB command prompt with an interface that lets scientists graphically construct molecular pathways by selecting species and reactions. SimBiology also includes a tabular interface for specifying reactants, products, parameters, and rules, as well as a number of capabilities needed by biologists, such as stochastic solvers, sensitivity analysis, and conservation of laws, including mass and energy.
SimBiology was designed to enable scientists and modelers to collaborate in the same software environment and complete their entire workflow with one tool. Biologists can build a model by graphically defining reactions. SimBiology then converts the defined reactions into a mathematical model that the modeler can refine, analyze, and simulate. In the same way, a modeler can create a complex, mathematically intensive model but leverage the graphical representation of the model to communicate their work with biologists.
Software tools have transformed engineering disciplines in the past decades, and they will play a vital role in helping systems biology reach its full potential. Evidence of this can already be seen in the number of major pharmaceutical companies that have transformed small proof-of-concept initiatives into fully funded Systems Biology departments.
What is Systems Biology?
Systems biology is a branch of computational biology that focuses on understanding how the biological world works at a system level. Systems biologists study the relationships between the components that make up an organism. Their goal is to develop accurate, unified models of biological activity—from the molecular level up to the entire organism—to enable the development of synthetic biological systems and accelerate drug discovery.
Closely related disciplines are bioinformatics, the development of algorithms and statistical techniques for the management and analysis of biological data, and PK/PD modeling, a technique used to model, simulate, and predict the effect of a drug on the body (pharmacodynamics) and the effect of the body on a drug (pharmacokinetics).
Published 2007 - 91483v00