rmabackadj
Perform background adjustment on Affymetrix microarray probe-level data using Robust Multi-array Average (RMA) procedure
Syntax
BackAdjustedMatrix = rmabackadj(PMData)
BackAdjustedMatrix =
rmabackadj(..., 'Method', MethodValue,
...)
BackAdjustedMatrix = rmabackadj(...,
'Truncate', TruncateValue, ...)
BackAdjustedMatrix = rmabackadj(...,
'Showplot', ShowplotValue, ...)
Input Arguments
PMData | Matrix of intensity values where each row corresponds to a perfect match (PM) probe and each column corresponds to an Affymetrix® CEL file. (Each CEL file is generated from a separate chip. All chips should be of the same type.) |
MethodValue | Specifies the estimation method for the background adjustment model parameters. Enter
either |
TruncateValue | Specifies the background noise model. Enter either |
ShowplotValue | Controls the plotting of a histogram showing the distribution
of PM probe intensity values (blue) and the convoluted probability
distribution function (red), with estimated parameters mu, sigma and
alpha. Enter either |
Output Arguments
BackAdjustedMatrix | Matrix of background-adjusted probe intensity values. |
Description
BackAdjustedMatrix = rmabackadj(PMData)PMData. Note that each row in PMData
corresponds to a perfect match (PM) probe and each column in PMData
corresponds to an Affymetrix CEL file. (Each CEL file is generated from a separate chip. All chips should be of
the same type.) Details on the background adjustment are described by Bolstad, 2005.
BackAdjustedMatrix = rmabackadj(...,
'PropertyName', PropertyValue, ...)rmabackadj with
optional properties that use property name/property value pairs. You
can specify one or more properties in any order. Each PropertyName must
be enclosed in single quotation marks and is case insensitive. These
property name/property value pairs are as follows:
specifies the estimation method for the background adjustment model parameters. When
BackAdjustedMatrix =
rmabackadj(..., 'Method', MethodValue,
...)MethodValue is 'RMA',
rmabackadj implements the estimation method described by Bolstad, 2005. When MethodValue is 'MLE',
rmabackadj estimates the parameters using maximum likelihood. Default is
'RMA'.
BackAdjustedMatrix = rmabackadj(...,
'Truncate', TruncateValue, ...)TruncateValue is false, rmabackadj uses
nontruncated Gaussian as the background noise model. Default is true.
BackAdjustedMatrix = rmabackadj(...,
'Showplot', ShowplotValue, ...)ShowplotValue is 'all', rmabackadj plots
a histogram for each column or chip. When ShowplotValue is
a number, list of numbers, or range of numbers, rmabackadj plots
a histogram for the indicated column number (chip).
For example:
(..., 'Showplot', 3,...)plots the intensity values in column 3 ofPMData.(..., 'Showplot', [3,5,7],...)plots the intensity values in columns 3, 5, and 7 ofPMData.(..., 'Showplot', 3:9,...)plots the intensity values in columns 3 to 9 ofPMData.
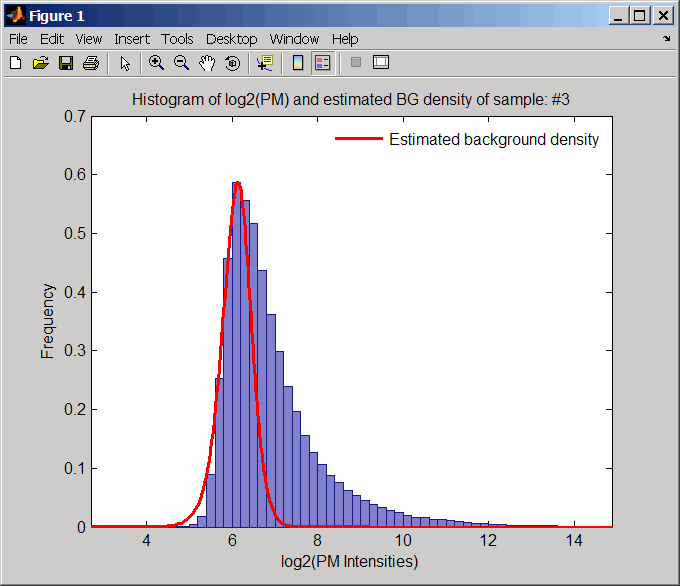
Examples
Load a MAT-file, included with the Bioinformatics Toolbox™ software, which contains Affymetrix probe-level data, including
pmMatrix, a matrix of PM probe intensity values from multiple CEL files.load prostatecancerrawdataPerform background adjustment on the PM probe intensity values in the matrix,
pmMatrix, creating a new matrix,BackgroundAdjustedMatrix.BackgroundAdjustedMatrix = rmabackadj(pmMatrix);
Perform background adjustment on the PM probe intensity values in only column 3 of the matrix,
pmMatrix, creating a new matrix,BackgroundAdjustedChip3.BackgroundAdjustedChip3 = rmabackadj(pmMatrix(:,3));
The prostatecancerrawdata.mat file used in
the previous example contains data from Best et al., 2005.
References
[1] Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., Speed, T.P. (2003). Exploration, Normalization, and Summaries of High Density Oligonucleotide Array Probe Level Data. Biostatistics 4, 249–264.
[2] Bolstad, B. (2005). “affy: Built-in Processing
Methods” https://www.bioconductor.org/packages/2.1/bioc/vignettes/affy/
inst/doc/builtinMethods.pdf
[3] Best, C.J.M., Gillespie, J.W., Yi, Y., Chandramouli, G.V.R., Perlmutter, M.A., Gathright, Y., Erickson, H.S., Georgevich, L., Tangrea, M.A., Duray, P.H., Gonzalez, S., Velasco, A., Linehan, W.M., Matusik, R.J., Price, D.K., Figg, W.D., Emmert-Buck, M.R., and Chuaqui, R.F. (2005). Molecular alterations in primary prostate cancer after androgen ablation therapy. Clinical Cancer Research 11, 6823–6834.
Version History
Introduced in R2006a
See Also
affyinvarsetnorm | affyread | affyrma | celintensityread | probelibraryinfo | probesetlookup | probesetvalues | quantilenorm | rmasummary
