Resultados de
- Allow MathWorks and the community leaders to easily post newsworthy items to the community
- Allow community visitors to respond to these posts with Likes and Replies
- Allow anyone to follow/subscribe the channel so they can be notified of new posts
- New or upcoming community features or events
- User highlights (e.g. examples of good behavior, interesting posts)
- Interesting content (e.g. File Exchange pick of the week submissions)
- Release notes and new features
- Polls (future)
The following is a list of updates and new features for MATLAB Central, including MATLAB Answers, File Exchange, Blogs, and Cody.
New Features
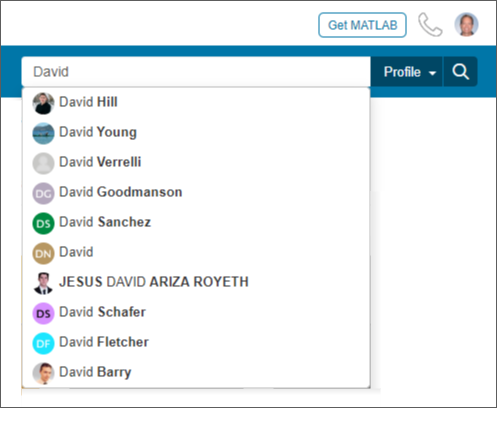
Profile search - A global community profile search has been added. The search field on community profile pages has been updated from a standard content search to a user profile search. This improvement makes it easier to find community members across all MATLAB Central. Previously one had to search the Answers contributors , File Exchange authors , and Cody players page when looking for a user profile.
Last seen - We have added a 'last seen' timestamp to community profiles which displays the date of a person's last visit to MATLAB Central. This can be a relevant bit of information and help determine how recent someone has been active in the community.
Answers pages design update - Answers Q&A pages have been updated to remove extra white space. This update includes smaller sized avatars, and position changes for the voting and content actions among other small changes. All these changes also help improve the mobile experience as well.
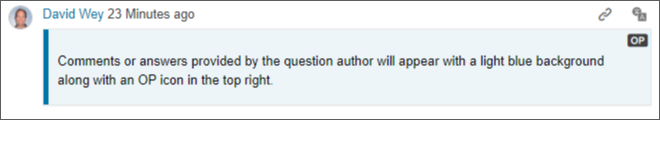
Original poster styles - Original poster styles have been introduced in Answers. When a question author participates in a Q&A thread their comments or answers will be styled with a blue background and left border so they're easily discernable from other contributors.
File Exchange data in monthly emails - File Exchange stats will be included in the monthly email we send to contributors who've participated in the community on any given month.
Trending content algorithm - The MATLAB Central home page trending content algorithm has been updated to look at content activity over a shorter period of time resulting in a more dynamic feed.
Walter Roberson does it again by winning the coveted MOST ACCEPTED answers badge for all his contributions in MATLAB Answers this past year. Walter has won this badge every year since 2015. It was way back in 2014 when Image Analyst out paced Walter and was awarded the badge.
There are 10 community members who have achieved the Top Downloads badge for their popular File Exchange submissions in 2019. Do you recognize any of these names? There's a good chance you've used one or more of their toolboxes or scripts in your work if you're a frequent visitor to File Exchange, if you're not you might want to check out what they've posted, it may save you a lot of time writing your own code.
--------------------- Top Downloads Badge Winners -----------------
- Diego Barragán
- Dirk-Jan Kroon
- Yi Cao
- John D'Errico
- Yair Altman
- Giampiero Campa
- Michael Kleder
- Dr. Siva Malla
- Antonio Trujillo-Ortiz
- Brett Shoelson
Congratulations to all these winners and a giant THANK YOU for all they've done this past year to help everyone in the MATLAB Central community!

We are happy to announce that virtual badges can now be achieved for participating in MATLAB Central File Exchange . We have 30 badges that anyone can achieve, which will also boost your community profile. Some badges are relatively simple to get while others will depend on how useful your submissions are to others in the community. Check out Ned Gulley's blog post for a great introduction.

Explore resources, ask questions, and discuss topics related to using Simulink to apply power electronics control to Electric Vehicles, Renewable Energy, Battery Systems, Power Conversion, and Motor Control. This is the 3rd MATLAB Central community, after Maker and SimBiology , and is moderated by Tony Lennon . Tony is the Power Electronics Marketing Manager at MathWorks.
Visit the community here . As always, let us know what you think by liking this post or commenting below.

I am fitting a generic TMDD model to date. Model fits look reasonable. Once I create a variant and simulate data for various doses to create observed vs predicted concentrations vs time profile, then simulated concentrations do not match fitted profiles. Simulated concentrations are either significantly higher or lower than the model fits. Not sure what is wrong. Fits look fine but simulated profiles using fitted parameters are all over the place.
Hello all,
I have created an arbitrary model for microtubule behavior. More or less just trying to familiarize with the software. I have created plots for multiple types of reactions that may occur and am looking to now plot the instantaneous derivative of each reaction. Would anyone have any suggestions as to how I could do this? I am familiar with how to do it with a clearly defined function with x,y,z,etc. values. However the 'sbiomodel' command doesn't seem to show me a function so I'm really lost on this.
Thanks for any and all help/suggestions.
Hello,
I have a suggestion for a feature which I think would be nice to add in SimBiology: a "date modified" field for all of the objects (species, parameters, rules, etc). It would be nice for this to be visible in the tables in the GUI, as well as being available through code, and to be able to sort on this field in the GUI. The reason I thought about this is because I just got a model with some updates from a colleague, and this would help me to identify what they updated.
Thank you,
Abed
In previous versions, there was an option for exporting fit results as a report. Now it is not there.
There will be an introductory hand-on SimBiology workshop following ACoP on Friday, Oct 24 at University of Florida, Orlando Campus, sponsored by Prof. Sihem Bihorel.
To register, please send me a direct message from the community site or e-mail me at fbuyukoz[at]mathworks.com. Please also let your colleagues know who might be interested in attending. Space is limited and will be allocated to those who sign up first.
Agenda
Introduction
- Overview of MATLAB and SimBiology
- Navigating the SimBiology desktop environment
- Working with a SimBiology project
Building and Simulating Mechanistic Models, using a TMDD example
- Overview of the building blocks and modeling architecture
- Building models using the SimBiology block diagram editor
- Configuring simulation-related settings (solvers, tolerances, sampling time, etc.)
- Exploring model dynamics and sensitivity using parameter sliders and sensitivity analysis
- Simulating hypothetical scenarios and dosing schedules
Implementing Traditional Compartmental PK/PD Workflows, using a 1 compartment PK model as example
- Importing, processing and visualizing data
- Performing non-compartmental analysis (NCA)
- Estimating parameters using nonlinear regression and population-based methods
Programmatic SimBiology and Integration with MATLAB
- Writing custom analysis tasks
- Automating workflows using MATLAB scripts
Over the past several months our communities have been the target of ongoing spam attacks. Spam is a common issue online and spam prevention is something we include as part of our standard development processes. The most recent attempts appear to be specifically targeting our sites with a combination of manual and automated attacks, probing with a variety of content attempting to bypass our filters. Regrettably, the attacks sometimes make it through our defenses, cluttering your inboxes and the content in our communities.
Rest assured, we are dedicated to continuing to evolve our tools and improve our capabilities to meet the goal of eliminating all spam in our communities. We appreciate your patience and understanding as we work to get there.
Sincerely,
David Wey
MATLAB Communities Development Manager
MathWorks, Inc.
ACoP10 Workshop – October 19, 2019
QSP Model Development Using gQSPSim: A GUI-Based Open-Source Platform for SimBiology Models
Organized by Genentech and MathWorks

IS there a quick and easy way to calculate AUC in SimBiology without exporting results to an NCA analysis>?
can we directly get the SD of the concentration-time AUC after fitting in Simbiology?
Hi, How do I get a numeric integral from a function that uses Excel file input data? I have been able to import the excel file but am having trouble continuing the code,please guide me, thank you
We have a Systems Biology and Biotechnology Specialization on Coursera which has the following specific courses:
1. Introduction to Systems Biology
2. Experimental Methods in Systems Biology
3. Network Analysis in Systems Biology
4. Dynamical Modeling Methods for Systems
5. Integrated Analysis in Systems Biology
6. Systems Biology and Biotechnology Capstone
For "Systems Biology and Biotechnology Capstone", "Dynamical Modeling Methods for Systems", and ":Integrated Analysis in Systems Biology", MATLAB is used for mathematical models and bioinformatics analyses. All assignments with dynamical models are also presented as MATLAB codes and SimBiology Models. The Systems Biology Specialization covers the concepts and methodologies used in systems-level analysis of biomedical systems. Successful participants will learn how to use experimental, computational and mathematical methods in systems biology and how to design practical systems-level frameworks to address questions in a variety of biomedical fields. In the final Capstone Project, students will apply the methods they learned in five courses of specialization to work on a research project.
Here is the link to all of these courses:
https://www.coursera.org/specializations/systems-biology
If you have any question about these courses please let me know.
Iman Tavassoly MD, PhD Icahn School of Medicine at Mount Sinai
Hi, I am intrigued by the idea of using the simbiology stochastic solvers for a project that I have so far coded in the idnlgrey framawork.
Some of the ODE right hand sides (ionic fluxes) in my model are given via fitobjects.
My question is: can I use a fit object in simbiology? Or should I figure out an analytical form and use it as a custom reaction rate?
Thanks, Francesco
On using Simbiology, I am realizing how wonderful it is! It is equivalent and infact better than much commercial software available in the market for hefty prices (not to name anyone in purpose). Moreover, the product is backed up by the world leaders in software engineering - MATLAB (which gives more confidence to the product). It would be very helpful if someone can share the list or some of the PubMed indexed publication on population pharmacokinetics in which Simbiology is utilized for modeling and computation.
Identification of model (one compartment, two compartments or three compartments) which a drug follows is an important step before population pharmacokinetic modeling. I am aware that the graph between the concentration vs time, gives an idea of the number of compartment a drug follows.
But is there a standard way to explore and determine the number of compartment a drug follows in a more objective manner. This would also be helpful to determine the model in which the data needs to be fit. In addition, a note on determining the order of reaction is also welcomed and would make the discussion complete.