Resultados de
This is not a question, it is my attempt at complying with the request for thumbs up/down voting. I vote thumbs up, for having AI.....
I am not sure if specific AI errors are to be reported. Other messages I just read from others here and the AI Chat itself clearly state that errors abound.
My AI request was: "Plot 300 points of field 2"
AI Chat gave me, in part:
data = thingSpeakRead(channelID, 'Fields', 2, 'NumPoints', 300, 'ReadKey', readAPIKey);
% Extract the field values
field1Values = data.Field1;
% Plot the data
plot(field1Values);
The AI code failed due to "Dot indexing is not supported for variables of this type"
So, I corrected the code thus to get the correct plot:
data = thingSpeakRead(channelID, 'Fields', 2, 'NumPoints', 300, 'ReadKey', readAPIKey);
% Extract the field values
%field1Values = data.Field1;
% Plot the data
plot(data);
I see great promise in AI Chat.
Opie
Hi! I'm new to pk modeling and Matlab. Can someone guide me through how to conduct population pk modeling based on pk parameters from non-human primate studies? Much thanks!!!!
Write a matlab script that will print the odd numbers, 1 through 20, in reverse.
I cannot figure out how to do this correctly, please help.
The title is resonably non-descript, but I can explain it easily:
Say I have an initial Emax model:
v = emax1*[G]^n1/(ec501^n1+[G]^n1)
And I want to place v inside of a second Emax model:
y = emax2*v^n2/(ex502^n2+v^n2)
Currently, I have the full function of v inside y, twice, it's very long and whilst I only need to get it correct once, for readability in the future I'd rather have it in form #2. I've played around with non-constant parameters but I need the steady state to be v, not the rate rule, and I haven't worked out how to make a parameter shift to a form like v, as an observation might.
Are there any recommended solutions or do I simply need to keep with having v fully expressed in y?
Thank you,
Dan
I saw this post on Answers.
I was impressed at the capability of the AI, as I have been at other times when I posed a question to it, at least some of the time. So much so that I wondered...
What if the AI were automatically applied to EVERY question on Answers? Would that be a good or bad thing? For example, suppose the AI automatically offers an answer to every question as soon as it gets posted? Of course, users would still be allowed to post their own, possibly better answers. But would it tend to disincentivise individuals from ansering questions?
Perhaps as bad, would it push Answers into the mode of a homework solving forum? Since if every homework question gets a possibly pretty good automatic AI generated solution, then every student will just post all HW questions, and the forum would quickly become overwhelmed.
I suppose one idea could be to set up the AI to post an answer to all un-answered questions that are at least one month old. Then students would not gain by posting their homework.
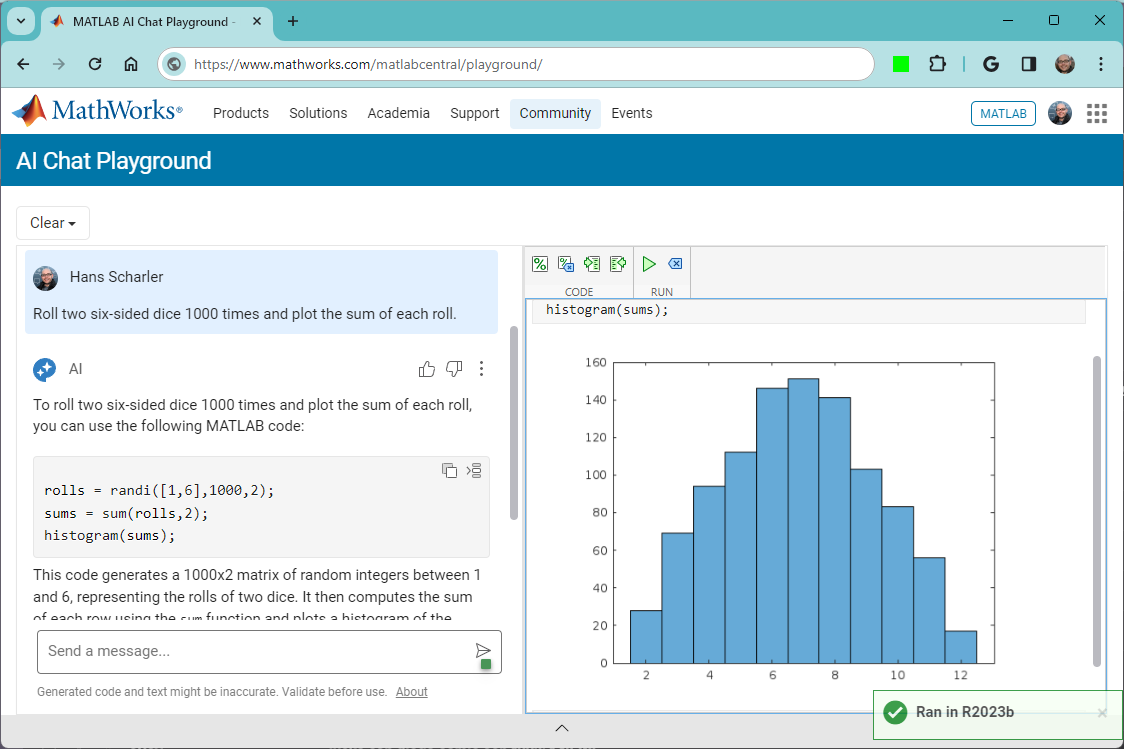
The MATLAB AI Chat Playground is open to everyone!
Check it out here on the community: https://www.mathworks.com/matlabcentral/playground
Hi All,
I'm currently attempting to implement a Hodgkin-Huxley-type model of membrane potential, ideally I would like a species that represents the membrane potential as its own distinct entity, so as the reference elsewhere. I've currently established a molarity-based work around but it would be great if I could set the units for the species as millivolt, but that throws an error.
Is there an established way to do this? I imagine I'm not the first person to be trying to model a voltage-gated ion channel!
Thank you for your help.
Hello,
I've looked around and I haven't found anything obvious about this, but is it possible to link to species/reactions, graphically, in a non-mass transfer sense? I have areas in my model where it would conceptually make sense to be able to see that species or reactions are linked, but if I link them in the standard way it demands that it be involved in the stoichiometry.
Perhaps some kind of dotted line, or similar?
Thank you, best regards,
Dan
Hello all,
I've been trying to shift my workflow more towards simbiology, it has a lot of very interesting features and it makes sense to try and do everything in one place if it works well..! Part of my hesitancy into this was some bad experiences handling units in the past, though this was almost certainly all out of my own ignorance, relatedly:
Getting onto my question.
In this model I have a species traveling around the body via blow flow, think a basic PBPK model. My species are picomolarities, if everything is already in concentrations, why is it necessary to initially divide by the compartment volume? i.e. 1/Pancreas below.

If my model dealt in molar quantities this would make a lot of sense, the division would represent the transition to concentrations. This, however, now necessitates my parameters be in units of liter/minute, which is actually correct, but I'd like clarification on why it's correct, ha!
Perhaps this is more of a modelling question than a simbiology question, but if there are answers I'd love to hear them. Thanks!
Hi All,
I'm attempting to put a set of simbiology global sensitivity analysis plots into my thesis and I'm running into some issues with the GSA plots. Firstly, the figures are very large, it would be quite beneficial to grab a set of the plots and arrange them myself, is there any documentation on how to mess around with the '1x1 Sobol' produced by sbiosobol? Or just GSA plots in general.
The second problem is that the results appear to be relative to the most sensitive parameter in that run. Is it recommended to have a resonably sensitive 'baseline' parameter in each run? I find it difficult to compare plots when a not so sensitive parameter is being recorded as near '1' for the whole run because it's being stacked against a set of very insensitive parameters. I.e. if i have multiple sets of GSAs due to a large model, how can I easily compare results? If I could do some single run through with every parameter that would be the ideal, I imagine, but then the default plot would be half a mile off the bottom of my screen, haha! Perhaps there is a solution to the first question that might help there?
Thank you for your help,
Dan
Hi all,
I've translated a model from another piece of software (monolix) into simbio programmatically to make use of your very easy global sensitivity analysis system.
It looks a little something like this, for a 'single' line example:
r1 = addreaction(model,'InsI -> InsP');
r1.ReactionRate = 'InsI*kip/vi'; %- is + panc
k1 = addkineticlaw(r1, 'Unknown');
Multiplied about 20 fold, as you can see I have included my volumes within the reaction rates myself (vi). The model functions perfectly and I have corrected the outputs at the end:
[time, x, names] = sbiosimulate(model,csObj,dObj1);
x(:,1) = x(:,1)/vi;
So that they are in concentration, as needed. However, when it comes to sensitivity analysis because I have corrected them post-model it is technically incorrect, it is analysing the absolute quantities. This is quite noticible in the sensitivity to the volumes.
Is there an easy fix to this, I've had to fight dimensionality with units in the past using simbio and I'd be great if there was some way of dividing a compartment output by a volume, for example. It is a functionality that exists in monolix, so I was hopeful it might here!
Thank you for your time.
EDIT:
I think I've worked it out, I had to refactor my model to operate in concentrations, just refitting it now. Now I should just be able to use unitless compartments.
I am trying to simulate model of blood lymphocyte count from a paper using Simbiology. The rate of in or out following circadian rythym is kp(t)=km + kb cos [(t-tpeak)*2pi/24] Where and how do I write the expression ? I dont think I can write in repeated assignment ?
I am currently facing a compatibility issue when attempting to load a SimBiology model created in MATLAB 2021a into MATLAB 2023a. Specifically, the code capture functionality does not seem to be working.
If anyone has encountered a similar situation or has insights on how to capture the code for a SimBiology model created in an older version of MATLAB, I would greatly appreciate your guidance. Are there any alternative methods or specific steps that can be followed to ensure successful code capture?
Thank you in advance for your time and expertise.
Dear community,
I would like to develop a fermentation model with 4 ODEs, one of which contains variable y. A "repeated assignment", e. g. y=5x+5, contains variable x that has been measured each second. These data (columns with time and corresponding value x in each row) are recorded in the Excel file.
Does anyone have any suggestion how to implement this in symbiology?
Thank you very much in advance,
Tetiana
Hi,
I want to develop a PK model based on some PK data. The PK data seems to display 2 peaks when one initial dose is given.
I would like to give one dose. A fraction of this dose (fr) is absorbed following the linear relationship - ka*Drug the other fraction (1-fr) is absorbed following a linear absorption (ka1*Drug) with a Tlag (it maybe a zero order). The fraction fr is unknown so it must be estimate.
Does anyone have can provide any suggestion to implement this in symbiology or provide a link where I can look?
Thank you very much in advance,
Ferran
I have a species which is absorbed in a zero-order manner. I want to model the reaction rate as "UptakeRate*ActiveState". So I can give value of 1 or 0 to ActivateState to control the active state of this process. The species itself is under control multiple processes, so I want to use Event to automatically change the active state of absorption.
However, when I use these two events:
Event 1,trigger: species > 0, Function: ActiveState =1; Event 2, trigger: species <=0, Function: ActiveState=0
The simulation seems to be stuck without stop.
Is there any way to solve this ? Thanks
Hi,
Im a new user of simbiology, can any one tell me How to fix a parameters to a known constant when estimating models.
Hello,
In the previous versions, we were able to change the reaction line properties to Reactant, Product, or Reactant and Product. This option is not available in 2021b. How do we change the proerpties if we reaction line is both Reactant and Product?
Thanks
Simulations and model results do not get exported to the MATLAB workspace automatically. 2021b. Under Model Analyzer preferences, I checked "export results when model run is complete", but to no effect. I have to manually expert simulations after each run. Please advise. Thanks RD