Entrenar una red con datos con valores complejos
En este ejemplo se muestra cómo predecir la frecuencia de una forma de onda de valores complejos mediante una red neuronal convolucional 1D.
Para pasar datos con valores complejos a una red neuronal, puede utilizar la capa de entrada para dividir los valores complejos en partes reales e imaginarias antes de que pase los datos a las capas posteriores de la red. Cuando la capa de entrada divide los datos de entrada de esta forma, la capa genera como salida los datos divididos como canales adicionales. Este diagrama muestra cómo fluyen los datos con valores complejos por una red neuronal convolucional.

Para dividir los datos con valores complejos en partes reales e imaginarias como su entrada a la red, establezca la opción SplitComplexInputs de la capa de entrada de la red en 1 (true).
En este ejemplo se entrena una red con regresión de secuencia a uno usando el conjunto de datos de forma de onda compleja, que contiene 500 formas de onda de valores complejos generadas de forma sintética de diferentes longitudes con dos canales. La red entrenada en este ejemplo predice la frecuencia de las formas de onda.
Cargar datos secuenciales
Cargue los datos de ejemplo de ComplexWaveformData.mat. Los datos son un arreglo de celdas de numObservations por 1 de secuencias, donde numObservations es el número de secuencias. Cada secuencia es un arreglo de valores complejos de numChannels por numTimeSteps, donde numChannels es el número de canales de la secuencia y numTimeSteps es el número de unidades de tiempo de la secuencia. Los objetivos correspondientes se encuentran en un arreglo numérico de numObservations por numResponses de las frecuencias de las formas de onda, donde numResponses es el número de canales de los objetivos.
load ComplexWaveformDataVisualice el número de observaciones.
numObservations = numel(data)
numObservations = 500
Visualice los tamaños de las primeras secuencias y las frecuencias correspondientes.
data(1:4)
ans=4×1 cell array
{2×157 double}
{2×112 double}
{2×102 double}
{2×146 double}
freq(1:4,:)
ans = 4×1
5.6232
2.1981
4.6921
4.5805
Visualice el número de canales de las secuencias. Para entrenar la red, cada secuencia debe tener el mismo número de canales.
numChannels = size(data{1},1)numChannels = 2
Visualice el número de respuestas (el número de canales de los objetivos).
numResponses = size(freq,2)
numResponses = 1
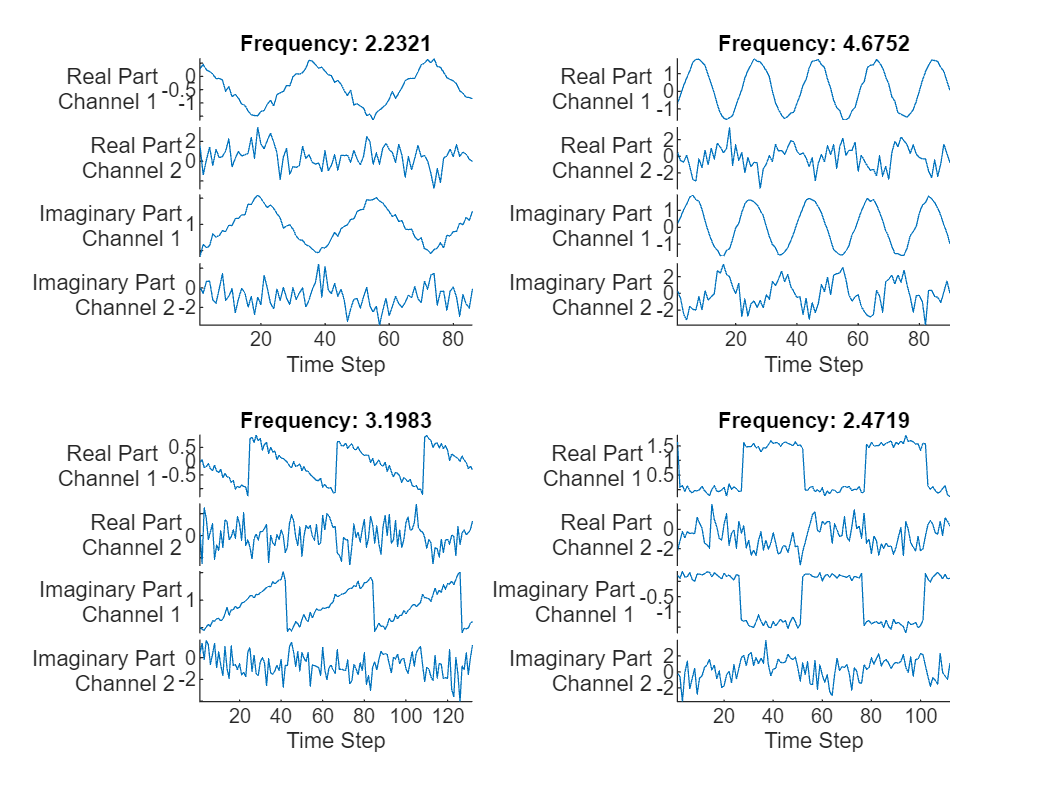
Visualice las primeras secuencias en gráficas.
displayLabels = [ ... "Real Part" + newline + "Channel " + string(1:numChannels), ... "Imaginary Part" + newline + "Channel " + string(1:numChannels)]; figure tiledlayout(2,2) for i = 1:4 nexttile stackedplot([real(data{i}') imag(data{i}')],DisplayLabels=displayLabels) xlabel("Time Step") title("Frequency: " + freq(i)) end

Preparar datos para el entrenamiento
Reserve datos para validación y pruebas. Divida los datos en un conjunto de entrenamiento que contenga el 80% de los datos, un conjunto de validación que contenga el 10% de los datos y un conjunto de prueba que contenga el 10% restante de los datos. Para dividir los datos, use la función trainingPartitions, incluida en este ejemplo como un archivo de soporte. Para acceder al archivo, abra el ejemplo como un script en vivo.
[idxTrain,idxValidation,idxTest] = trainingPartitions(numObservations, [0.8 0.1 0.1]); XTrain = data(idxTrain); XValidation = data(idxValidation); XTest = data(idxTest); TTrain = freq(idxTrain); TValidation = freq(idxValidation); TTest = freq(idxTest);
Para ayudar a comprobar que la red es válida para las secuencias de entrenamiento más cortas, puede pasar la longitud de la secuencia más corta a la capa de entrada de secuencias de la red. Calcule la longitud de la secuencia de entrenamiento más corta.
for n = 1:numel(XTrain) sequenceLengths(n) = size(XTrain{n},2); end minLength = min(sequenceLengths)
minLength = 76
Definir la arquitectura de la red convolucional 1D
Defina la arquitectura de la red neuronal convolucional 1D.
Especifique una capa de entrada de secuencias con un tamaño de entrada que coincida con el número de características de los datos de entrada.
Para dividir los datos de entrada en partes reales e imaginarias, establezca la opción
SplitComplexInputsde la capa de entrada en1(true).Para ayudar a comprobar que la red es válida para las secuencias de entrenamiento más cortas, establezca la opción
MinLengthen la longitud de la secuencia de entrenamiento más corta.Especifique dos bloques de capas convolucionales 1D, ReLU y normalización de capas, donde la capa convolucional tiene un tamaño de filtro de 5. Especifique 32 y 64 filtros para la primera y la segunda capa convolucional, respectivamente. Para ambas capas convolucionales, rellene la parte izquierda de las entradas de manera que las salidas tengan la misma longitud (causal padding).
Para reducir la salida de las capas convolucionales a un solo vector, utilice una capa de agrupación media global 1D.
Para especificar el número de valores que se desea predecir, incluya una capa totalmente conectada con un tamaño que coincida con el número de respuestas.
filterSize = 5; numFilters = 32; layers = [ ... sequenceInputLayer(numChannels,SplitComplexInputs=true,MinLength=minLength) convolution1dLayer(filterSize,numFilters,Padding="causal") reluLayer layerNormalizationLayer convolution1dLayer(filterSize,2*numFilters,Padding="causal") reluLayer layerNormalizationLayer globalAveragePooling1dLayer fullyConnectedLayer(numResponses)];
Especificar las opciones de entrenamiento
Especifique las opciones de entrenamiento.
Entrenar usando el optimizador Adam.
Especificar que los datos de entrada tienen formato
"CTB"(canal, tiempo, lote).Entrenar durante 250 épocas. Para conjuntos de datos más grandes, puede que no sea necesario entrenar durante tantas épocas para conseguir un buen ajuste.
Especificar las secuencias y las respuestas usadas para la validación.
Generar la red que dé la pérdida de validación más baja.
Mostrar el proceso de entrenamiento en una gráfica.
Deshabilitar la salida detallada.
options = trainingOptions("adam", ... InputDataFormats="CTB", ... MaxEpochs=250, ... ValidationData={XValidation, TValidation}, ... OutputNetwork="best-validation-loss", ... Plots="training-progress", ... Verbose=false);
Entrenar la red
Entrene la red neuronal con la función trainnet. Para la regresión, utilice la pérdida de error cuadrático medio. De forma predeterminada, la función trainnet usa una GPU en caso de que esté disponible. Para utilizar una GPU se requiere una licencia de Parallel Computing Toolbox™ y un dispositivo GPU compatible. Para obtener información sobre los dispositivos compatibles, consulte GPU Computing Requirements (Parallel Computing Toolbox). De lo contrario, la función usa la CPU. Para especificar el entorno de ejecución, utilice la opción de entrenamiento ExecutionEnvironment.
net = trainnet(XTrain,TTrain,layers,"mse",options);
Probar la red
Realice predicciones con los datos de prueba mediante la función minibatchpredict. De forma predeterminada, la función minibatchpredict usa una GPU en caso de que esté disponible. Rellene a la izquierda los datos y especifique que tienen formato "CTB" (canal, tiempo, lote).
YTest = minibatchpredict(net,XTest, ... SequencePaddingDirection="left", ... InputDataFormats="CTB");
Visualice las primeras predicciones en una gráfica.
displayLabels = [ ... "Real Part" + newline + "Channel " + string(1:numChannels), ... "Imaginary Part" + newline + "Channel " + string(1:numChannels)]; figure tiledlayout(2,2) for i = 1:4 nexttile stackedplot([real(XTest{i}') imag(XTest{i}')], DisplayLabels=displayLabels); xlabel("Time Step") title("Frequency: " + YTest(i)) end
Visualice los errores cuadráticos medios en un histograma.
figure histogram(mean((TTest - YTest).^2,2)) xlabel("Error") ylabel("Number of Predictions")

Calcule el error cuadrático medio raíz global.
rmse = sqrt(mean((YTest-TTest).^2))
rmse = single
0.6751
Represente las frecuencias predichas frente a las frecuencias objetivo.
figure scatter(YTest,TTest,"+"); xlabel("Predicted Frequency") ylabel("Target Frequency") hold on m = min(freq); M = max(freq); xlim([m M]) ylim([m M]) plot([m M], [m M], "--")

Consulte también
trainnet | trainingOptions | dlnetwork | testnet | minibatchpredict | predict | scores2label | convolution1dLayer | sequenceInputLayer