Breast Tumor Segmentation from Ultrasound Using Deep Learning
This example shows how to perform semantic segmentation of breast tumors from 2-D ultrasound images using a deep neural network.
Semantic segmentation involves assigning a class to each pixel in a 2-D image. In this example, you perform breast tumor segmentation using the DeepLab v3+ architecture. A common challenge of medical image segmentation is class imbalance. In segmentation, class imbalance means the size of the region of interest, such as a tumor, is small relative to the image background, resulting in many more pixels in the background class. This example addresses class imbalance by using a custom Tversky loss [1]. The Tversky loss is an asymmetric similarity measure that is a generalization of the Dice index and the Jaccard index.
Load Pretrained Network
Create a folder in which to store the pretrained network and image data set. In this example, a folder named BreastSegmentation created within the tempdir directory has been used as dataDir. Download the pretrained DeepLab v3+ network and test image by using the downloadTrainedNetwork helper function. The helper function is attached to this example as a supporting file. You can use the pretrained network to run the example without waiting for training to complete.
dataDir = fullfile(tempdir,"BreastSegmentation"); if ~exist(dataDir,"dir") mkdir(dataDir) end pretrainedNetwork_url = "https://www.mathworks.com/supportfiles/"+ ... "image/data/breast_seg_deepLabV3_v2.zip"; downloadTrainedNetwork(pretrainedNetwork_url,dataDir); load(fullfile(dataDir,"breast_seg_deepLabV3_v2.mat"));
Read the test ultrasound image and resize the image to the input size of the pretrained network.
imTest = imread(fullfile(dataDir,"breastUltrasoundImg.png"));
imSize = [256 256];
imTest = imresize(imTest,imSize);Predict the tumor segmentation mask for the test image. Specify the classes to predict as "tumor" and "background".
classNames = ["tumor","background"]; segmentedImg = semanticseg(imTest,trainedNet,Classes=classNames);
Display the test image and the test image with the predicted tumor label overlay as a montage.
overlayImg = labeloverlay(imTest,segmentedImg,Transparency=0.7,... IncludedLabels="tumor", ... Colormap="hsv"); montage({imTest,overlayImg});

Download Data Set
This example uses the Breast Ultrasound Images (BUSI) data set [2]. The BUSI data set contains 2-D ultrasound images stored in the PNG file format. The total size of the data set is 197 MB. The data set contains 133 normal scans, 487 scans with benign tumors, and 210 scans with malignant tumors. This example uses images from the tumor groups only. Each ultrasound image has a corresponding tumor mask image. The tumor mask labels have been reviewed by clinical radiologists [2].
Run this code to download the dataset from the MathWorks® website and unzip the downloaded folder.
zipFile = matlab.internal.examples.downloadSupportFile("image","data/Dataset_BUSI.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
The imageDir folder contains the downloaded and unzipped dataset.
imageDir = fullfile(filepath,"Dataset_BUSI_with_GT");Load Data
Create an imageDatastore object to read and manage the ultrasound image data. Label each image as normal, benign, or malignant according to the name of its folder.
imds = imageDatastore(imageDir,IncludeSubfolders=true,LabelSource="foldernames");Remove files whose names contain "mask" to remove label images from the datastore. The image datastore now contains only the grayscale ultrasound images.
imds = subset(imds,find(~contains(imds.Files,"mask")));Create a pixelLabelDatastore (Computer Vision Toolbox) object to store the labels. Specify the same class names as defined in the previous section. The pixel label ID 1 maps to the "tumor" class name, and the pixel label ID 0 maps to the "background" class name.
disp(classNames)
"tumor" "background"
labelIDs = [1 0]; numClasses = numel(classNames); pxds = pixelLabelDatastore(imageDir,classNames,labelIDs,IncludeSubfolders=true);
Include only the subset of files whose names contain "_mask.png" in the datastore. The pixel label datastore now contains only the tumor mask images.
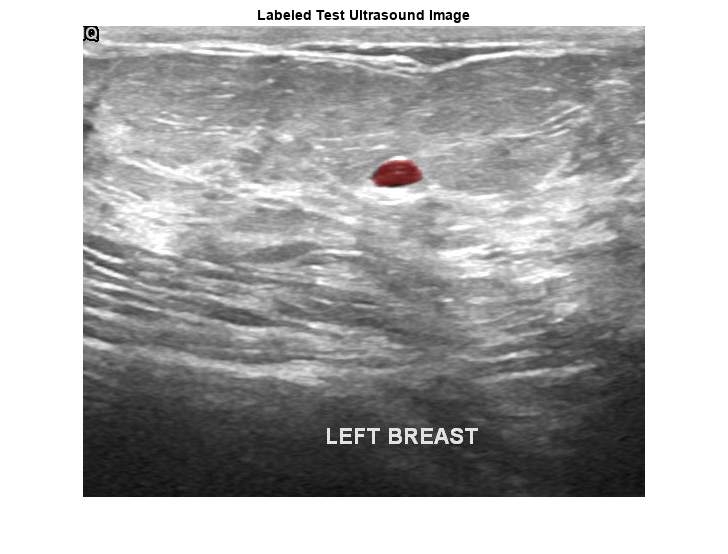
pxds = subset(pxds,contains(pxds.Files,"_mask.png"));Preview one image with a tumor mask overlay.
testImage = preview(imds); mask = preview(pxds); B = labeloverlay(testImage,mask,Transparency=0.7, ... IncludedLabels="tumor", ... Colormap="hsv"); imshow(B) title("Labeled Test Ultrasound Image")
Combine the image datastore and the pixel label datastore to create a CombinedDatastore object.
dsCombined = combine(imds,pxds);
Prepare Data for Training
Partition Data into Training, Validation, and Test Sets
Split the combined datastore into data sets for training, validation, and testing. Allocate 80% of the data for training, 10% for validation, and the remaining 10% for testing. Determine the indices to include in each set by using the splitlabels (Computer Vision Toolbox) function. To exclude images in the normal class without tumor images, use the image datastore labels as input and set the Exclude name-value argument to "normal".
idxSet = splitlabels(imds.Labels,[0.8,0.1],"randomized",Exclude="normal"); dsTrain = subset(dsCombined,idxSet{1}); dsVal = subset(dsCombined,idxSet{2}); dsTest = subset(dsCombined,idxSet{3});
Augment Training and Validation Data
Augment the training and validation data by using the transform function with custom preprocessing operations specified by the transformBreastTumorImageAndLabels helper function. The helper function is attached to the example as a supporting file. The transformBreastTumorImageAndLabels function performs these operations:
Convert the ultrasound images from RGB to grayscale.
Augment the intensity of the grayscale images by using the
jitterIntensityfunction.Resize the images to 256-by-256 pixels.
tdsTrain = transform(dsTrain,@transformBreastTumorImageAndLabels,IncludeInfo=true); tdsVal = transform(dsVal,@transformBreastTumorImageAndLabels,IncludeInfo=true);
Define Network Architecture
This example uses the DeepLab v3+ network. DeepLab v3+ consists of a series of convolution layers with a skip connection, one maxpool layer, and one averagepool layer. The network also has a batch normalization layer before each ReLU layer.
Create a DeepLab v3+ network based on ResNet-50 by using the using deeplabv3plus (Computer Vision Toolbox) function. Setting the base network as ResNet-50 requires the Deep Learning Toolbox™ Model for ResNet-50 Network support package. If this support package is not installed, then the function provides a download link.
Define the input size of the network as 256-by-256-by-3. Specify the number of classes as two for background and tumor.
imageSize = [256 256 3];
net = deeplabv3plus(imageSize,numClasses,"resnet50");Because the preprocessed ultrasound images are grayscale, replace the original input layer with a 256-by-256 input layer.
newInputLayer = imageInputLayer(imageSize(1:2),Name="newInputLayer");
net = replaceLayer(net,net.Layers(1).Name,newInputLayer);Replace the first 2-D convolution layer with a new 2-D convolution layer to match the size of the new input layer.
newConvLayer = convolution2dLayer([7 7],64,Stride=2,Padding=[3 3 3 3],Name="newConv1");
net = replaceLayer(net,net.Layers(2).Name,newConvLayer);Alternatively, you can modify the DeepLab v3+ network by using the Deep Network Designer (Deep Learning Toolbox) from Deep Learning Toolbox.
Use the Deep Network Designer (Deep Learning Toolbox) to analyze the DeepLab v3+ network.
deepNetworkDesigner(net)
Specify Training Options
Train the network using the adam optimization solver. Specify the hyperparameter settings using the trainingOptions (Deep Learning Toolbox) function. Set the learning rate to 1e-3 over the span of training. You can experiment with the mini-batch size based on your GPU memory. Batch normalization layers are less effective for smaller values of the mini-batch size. Tune the initial learning rate based on the mini-batch size.
options = trainingOptions("adam", ... ExecutionEnvironment="gpu", ... InitialLearnRate=1e-3, ... ValidationData=tdsVal, ... MaxEpochs=300, ... MiniBatchSize=16, ... Verbose=false, ... VerboseFrequency=20, ... Plots="training-progress");
Train Network
To train the network, set the doTraining variable to true. Train the model using the trainnet (Deep Learning Toolbox) function. To address the class imbalance between the smaller tumor regions and larger background, specify a custom loss function, tverskyLoss, which is defined as a helper function at the end of this example.
Train on a GPU if one is available. Using a GPU requires Parallel Computing Toolbox™ and a CUDA® enabled NVIDIA® GPU. For more information, see GPU Computing Requirements (Parallel Computing Toolbox). Training takes about four hours on a single-GPU system with an NVIDIA™ Titan Xp GPU and can take longer depending on your GPU hardware.
doTraining =false; if doTraining trainedNet = trainnet(tdsTrain,net,@tverskyLoss,options); modelDateTime = string(datetime("now",Format="yyyy-MM-dd-HH-mm-ss")); save("breastTumorDeepLabv3-"+modelDateTime+".mat","trainedNet"); end
Predict Using New Data
Preprocess Test Data
Prepare the test data by using the transform function with custom preprocessing operations specified by the transformBreastTumorImageResize helper function. This helper function is attached to the example as a supporting file. The transformBreastTumorImageResize function converts images from RGB to grayscale and resizes the images to 256-by-256 pixels.
dsTest = transform(dsTest,@transformBreastTumorImageResize,IncludeInfo=true);
Segment Test Data
Use the trained network for semantic segmentation of the test data set. Specify the same class names to predict as defined earlier in the example.
pxdsResults = semanticseg(dsTest,trainedNet,Verbose=true,Classes=classNames);
Running semantic segmentation network ------------------------------------- * Processed 65 images.
Evaluate Segmentation Accuracy
Evaluate the network-predicted segmentation results against the ground truth pixel label tumor masks.
metrics = evaluateSemanticSegmentation(pxdsResults,dsTest,Verbose=true);
Evaluating semantic segmentation results
----------------------------------------
* Selected metrics: global accuracy, class accuracy, IoU, weighted IoU, BF score.
* Processed 65 images.
* Finalizing... Done.
* Data set metrics:
GlobalAccuracy MeanAccuracy MeanIoU WeightedIoU MeanBFScore
______________ ____________ _______ ___________ ___________
0.96901 0.94712 0.84813 0.94467 0.59684
Measure the segmentation accuracy using the evaluateBreastTumorDiceAccuracy helper function. This helper function computes the Dice index between the predicted and ground truth segmentations using the dice function. The helper function is attached to the example as a supporting file.
[diceTumor,diceBackground,numTestImgs] = evaluateBreastTumorDiceAccuracy(pxdsResults,dsTest);
Calculate the average Dice index across the set of test images.
disp("Average Dice score of background across "+num2str(numTestImgs)+ ... " test images = "+num2str(mean(diceBackground)))
Average Dice score of background across 65 test images = 0.98047
disp("Average Dice score of tumor across "+num2str(numTestImgs)+ ... " test images = "+num2str(mean(diceTumor)))
Average Dice score of tumor across 65 test images = 0.81332
disp("Median Dice score of tumor across "+num2str(numTestImgs)+ ... " test images = "+num2str(median(diceTumor)))
Median Dice score of tumor across 65 test images = 0.85071
Visualize statistics about the Dice scores as a box chart. The middle blue line in the plot shows the median Dice index. The upper and lower bounds of the blue box indicate the 25th and 75th percentiles, respectively. Black whiskers extend to the most extreme data points that are not outliers.
figure boxchart([diceTumor diceBackground]) title("Test Set Dice Accuracy") xticklabels(classNames) ylabel("Dice Coefficient")

Supporting Functions
The tverskyLoss helper function specifies a custom loss function based on the Tversky loss metric. For more details about Tversky loss, see generalizedDice (Computer Vision Toolbox). The alpha and beta weighting factors control the contribution of false positives and false negatives, respectively, to the loss function. The alpha and beta values used in this example were selected using trial and error for the target data set. Generally, specifying the beta value greater than the alpha value is useful for training images with small objects and large background regions.
function loss = tverskyLoss(Y,T) % Copyright 2024 The MathWorks, Inc. % Specify weights alpha = 0.01; beta = 0.99; % Define a small constant to prevent division by zero. epsilon = 1e-8; % Compute the Tversky loss. Pcnot = 1-Y; Gcnot = 1-T; TP = sum(sum(Y.*T,1),2); FP = sum(sum(Y.*Gcnot,1),2); FN = sum(sum(Pcnot.*T,1),2); numer = TP + epsilon; denom = TP + alpha*FP + beta*FN + epsilon; lossTIc = 1 - numer./denom; lossTI = sum(lossTIc,3); % Compute the average Tversky loss. N = size(Y,4); loss = sum(lossTI)/N; end
References
[1] Salehi, Seyed Sadegh Mohseni, Deniz Erdogmus, and Ali Gholipour. “Tversky Loss Function for Image Segmentation Using 3D Fully Convolutional Deep Networks.” In Machine Learning in Medical Imaging, edited by Qian Wang, Yinghuan Shi, Heung-Il Suk, and Kenji Suzuki, 10541:379–87. Cham: Springer International Publishing, 2017. https://doi.org/10.1007/978-3-319-67389-9_44.
[2] Al-Dhabyani, Walid, Mohammed Gomaa, Hussien Khaled, and Aly Fahmy. “Dataset of Breast Ultrasound Images.” Data in Brief 28 (February 2020): 104863. https://doi.org/10.1016/j.dib.2019.104863.
See Also
imageDatastore | pixelLabelDatastore (Computer Vision Toolbox) | subset | combine | transform | deeplabv3plus (Computer Vision Toolbox) | semanticseg (Computer Vision Toolbox) | evaluateSemanticSegmentation (Computer Vision Toolbox)