Get Started with Medical Imaging
Medical imaging refers to the non-invasive acquisition and processing of images of the human body for clinical applications. Medical imaging often involves collaboration between clinicians, radiologists, and image processing experts. You can use medical image processing for applications including clinical diagnosis, surgical planning, and research.
This list provides some sample applications you can explore in Medical Imaging Toolbox™.
Diagnostic Systems: Detect tumors in brain MRI scans or breast ultrasound images, COVID-19 in CT scans, or pneumonia in chest X-rays.
Biomedical Engineering: Generate 3-D models of bones for finite element modeling, 3-D printing, or orthopedic implant design.
Functional Analysis: Analyze brain function from functional MRI, or estimate respiratory motion from CT scans.
Pharmaceutical Research: Measure drug efficacy and clearance time.
Device Design: Develop next generation imaging devices and algorithms.
Medical Imaging Modalities
A medical imaging modality refers to the technology used to capture the image, such as X-ray, magnetic resonance imaging (MRI), or computed tomography (CT). Medical Imaging Toolbox supports most 2-D and 3-D modalities including, but not limited to, CT, MRI, positron emission tomography (PET), single-photon emission CT (SPECT), ultrasound, X-ray, X-ray angiography, mammography, and cine-MRI. You can also use Medical Imaging Toolbox to explore multimodal data sets, which combine multiple modalities. Additionally, the Medical Imaging Toolbox Interface for Cellpose Library support package enables cell segmentation in microscopy images.
Note
Medical Imaging Toolbox functionality generally works with all images in supported medical file formats, regardless of modality.
This table lists a subset of common imaging modalities. Each entry provides basic background information, a representative image, tips for getting started, and links to examples that use the corresponding modality.
| Modality | Image | Get Started | See Also |
|---|---|---|---|
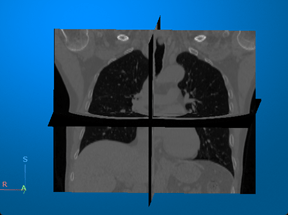
Computed tomography (CT) — Visualize 3-D patient anatomy using X-rays. |
For an example that uses this image [1], see Display Labeled Medical Image Volume in Patient Coordinate System. |
| |
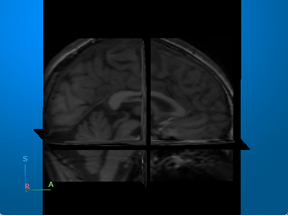
Magnetic resonance imaging (MRI) — Visualize 3-D patient anatomy using a powerful magnetic field and radio waves. |
For an example that uses this image [2][3], see Brain MRI Segmentation Using Pretrained 3-D U-Net Network. |
| |
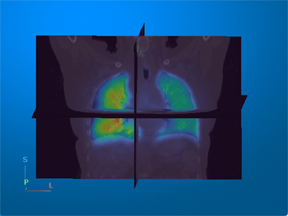
Positron emission tomography (PET) — Visualize metabolic or functional activity in 3-D using radioactive tracers. |
For an example that uses this image [4], see Display Multimodal Medical Image Data from PET and CT. |
| |
Ultrasound — Visualize 2-D patient anatomy using sound waves. |
For an example that uses this image, see Read, Process, and View Ultrasound Data. |
| |
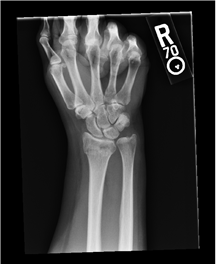
X-ray — Visualize 2-D anatomy using X-rays. |
For an example that uses this image, see Create Medical Image Object from Filename. |
| |
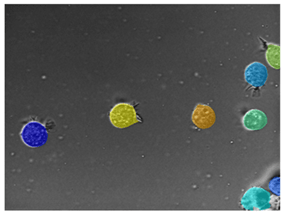
Microscopy — Visualize tissue samples or cells at the microscopic level. |
For an example that uses this image, see Getting Started with Cellpose. |
|
Typical Workflow for Medical Image Analysis
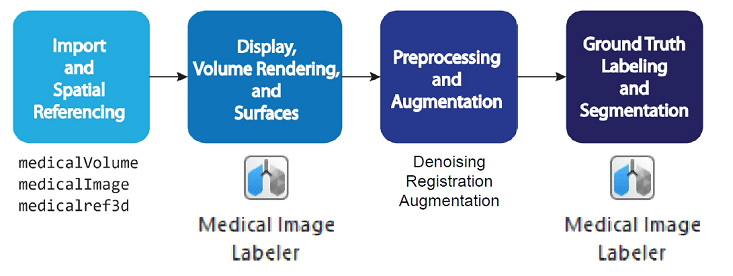
Import and Spatial Referencing
A typical medical imaging workflow begins with importing data into the workspace. Medical images are available in file formats such as NIfTI, DICOM, NRRD, Analyze7.5, and Interfile. Medical Imaging Toolbox provides general 2-D and 3-D medical image objects, as well as file-format-specific functions, that you can use to import and export medical images. For more information, see Read, Process, and Write 3-D Medical Images.
Display, Volume Rendering, and Surfaces
Once you have imported a medical image into the workspace, you can view and inspect the image to plan your workflow. Medical Imaging Toolbox provides tools for 2-D and 3-D display, and for generating surfaces for 3-D printing or modeling applications. For more information, see Choose Approach for Medical Image Visualization.
Preprocessing and Augmentation
Image preprocessing and augmentation prepare images for advanced workflows. Use image preprocessing to reduce artifacts such as noise, or to register multiple images. For deep learning workflows, you can increase the size of your training data set using augmentation, which applies random transformations to simulate variations in image acquisition and patient anatomy. For more information, see Medical Image Preprocessing.
Ground Truth Labeling, Segmentation, and Analysis
The toolbox supports interactive and deep learning-based image segmentation and pixel labeling using the Medical Image Labeler app. Label large ground truth data sets for training deep learning networks, or segment individual images to isolate regions of interest for analysis workflows. You can analyze segmented images using standardized radiomics features, or develop custom analysis workflows using image processing functions. For more information, see Get Started with Medical Image Labeler and Analysis and Applications.
References
[1] Medical Segmentation Decathlon. "Lung." Tasks. Accessed May 10, 2018. http://medicaldecathlon.com/. The Medical Segmentation Decathlon data set is provided under the CC-BY-SA 4.0 license. All warranties and representations are disclaimed. See the license for details.
[2] “NITRC: CANDI Share: Schizophrenia Bulletin 2008: Tool/Resource Info.” Accessed October 17, 2022. https://www.nitrc.org/projects/cs_schizbull08/.
[3] Frazier, J. A., S. M. Hodge, J. L. Breeze, A. J. Giuliano, J. E. Terry, C. M. Moore, D. N. Kennedy, et al. “Diagnostic and Sex Effects on Limbic Volumes in Early-Onset Bipolar Disorder and Schizophrenia.” Schizophrenia Bulletin 34, no. 1 (October 27, 2007): 37–46. https://doi.org/10.1093/schbul/sbm120.
[4] Eslick, Enid M., John Kipritidis, Denis Gradinscak, Mark J. Stevens, Dale L. Bailey, Benjamin Harris, Jeremy T. Booth, and Paul J. Keall. “CT Ventilation as a Functional Imaging Modality for Lung Cancer Radiotherapy (CT-vs-PET-Ventilation-Imaging).” The Cancer Imaging Archive, 2022. https://doi.org/10.7937/3PPX-7S22.
See Also
Medical Image
Labeler | medicalVolume | medicalImage